Learning Objectives
From today’s class, students are anticipated to be able to:
- Recognize how to manipulate data through a variety of tibble joins such as:
- Mutating joins:
left_join(),right_join(),full_join(),anti_join() - Filtering joins:
semi_join(),anti_join()
- Mutating joins:
- Perform binding:
bind_rows(),bind_cols() - Join more than 2 tibbles
- Join based on multiple conditions
- Perform set operations on data:
intersect(),union(),setdiff() - Join tibbles with different types of variables
Resources
Video lecture:
Demonstration .Rmd file:
Other resources, in addition to the notes below:
- Two-table verbs vignette
- Jenny’s Join Cheatsheet for a quick reference to joins.
- Relational Data chapter in “R for Data Science”.
- dplyr cheatsheet
Overview of join functions
Note: In order to merge two tibbles, you need to have an identifier variable that has unique values for every row of observations in both tibbles.
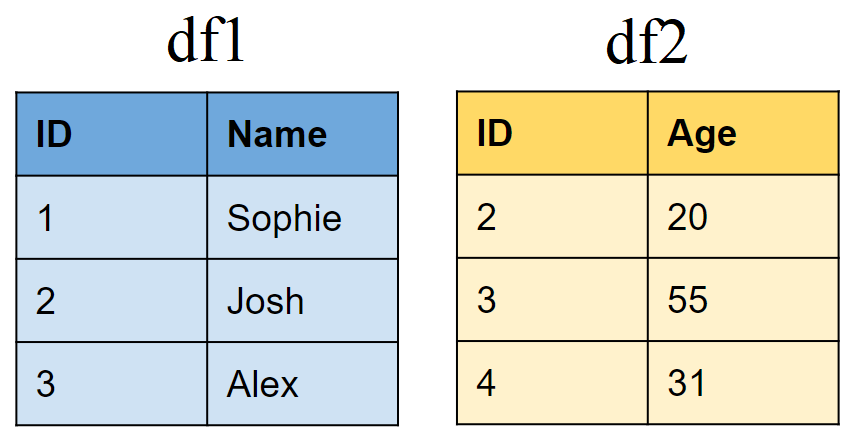
Create two sample tibbles:
# First tibble
df1 <- tibble(ID = 1:3,
Name = c("Sophie", "Josh","Alex"))
# Second tibble
df2 <- tibble(ID = 2:4,
Age = c(20,50,31))
Mutating joins
Join matching rows from df2 to df1
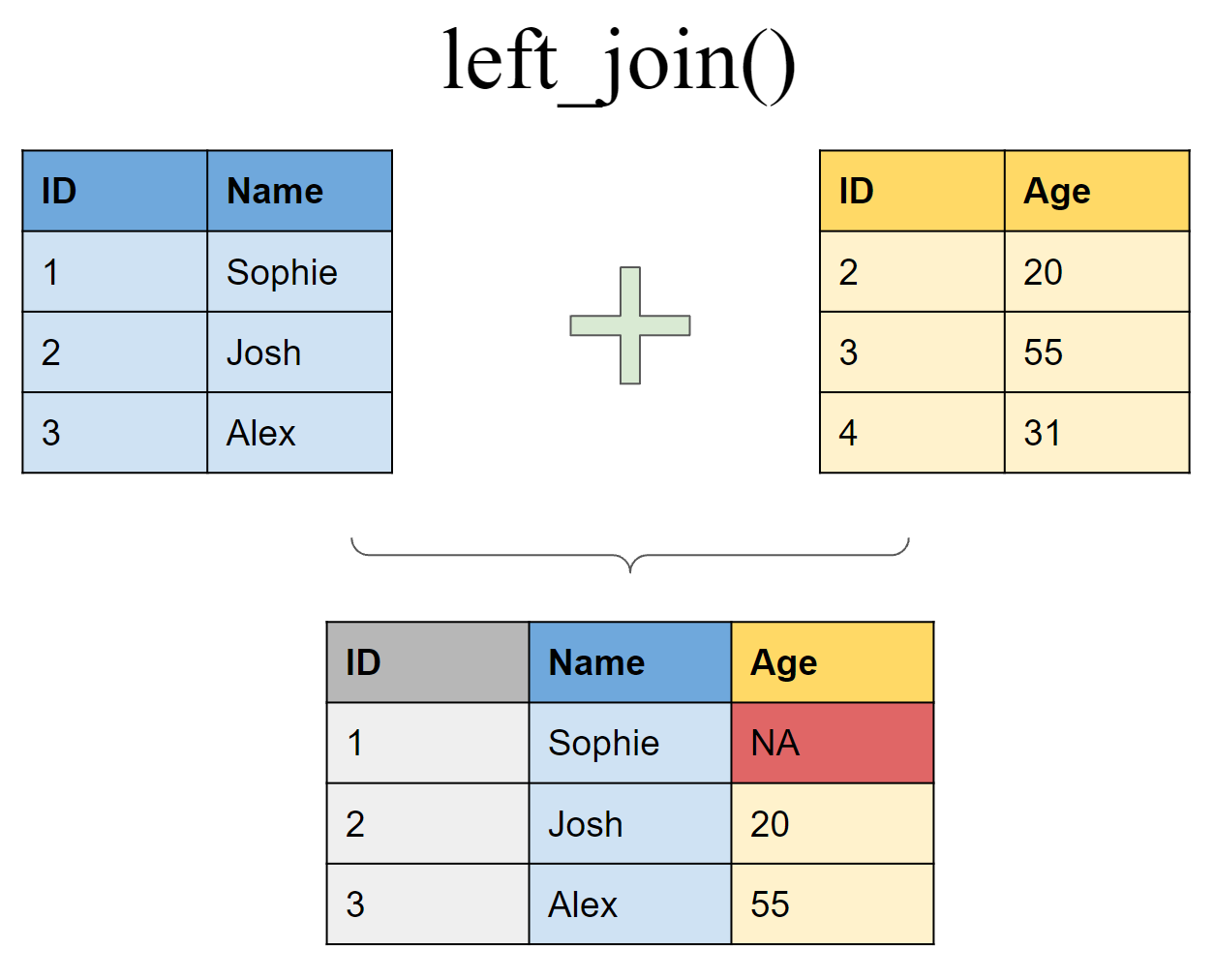
left_join(df1, df2, by = "ID")
## # A tibble: 3 x 3
## ID Name Age
## <int> <chr> <dbl>
## 1 1 Sophie NA
## 2 2 Josh 20
## 3 3 Alex 50
Join matching rows from df1 to df2
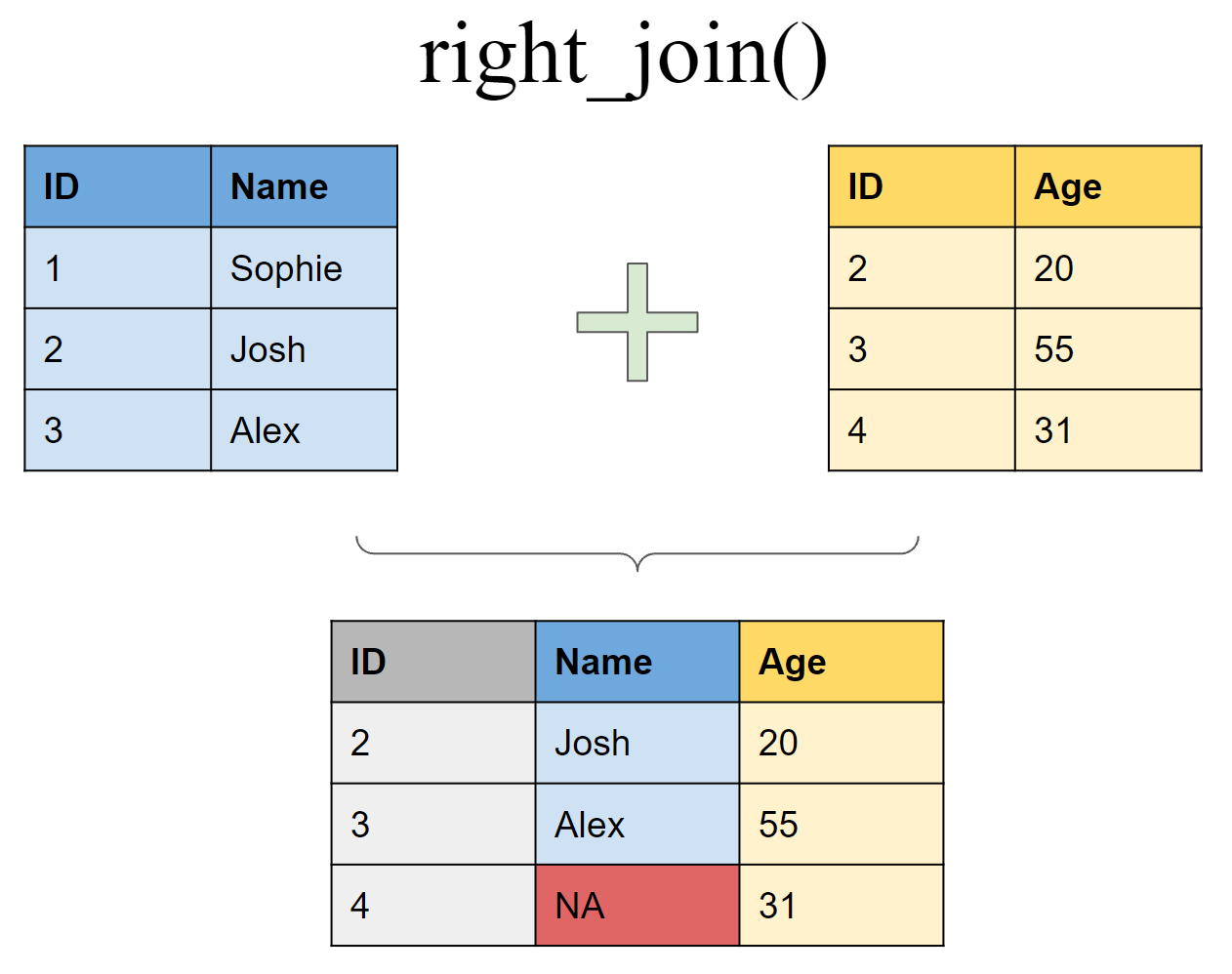
right_join(df1, df2, by = "ID")
## # A tibble: 3 x 3
## ID Name Age
## <int> <chr> <dbl>
## 1 2 Josh 20
## 2 3 Alex 50
## 3 4 <NA> 31
Retain only rows present in both sets
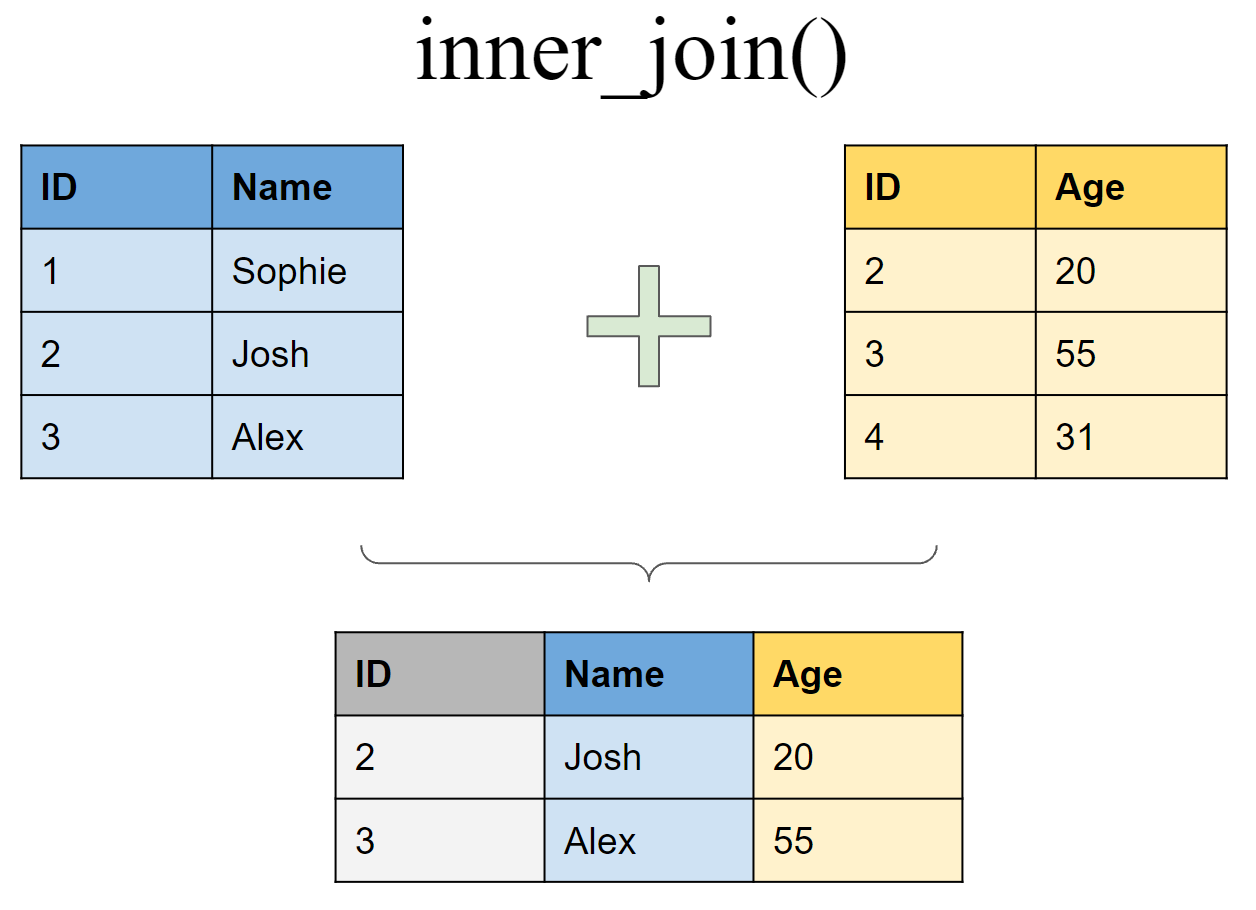
inner_join(df1, df2, by = "ID")
## # A tibble: 2 x 3
## ID Name Age
## <int> <chr> <dbl>
## 1 2 Josh 20
## 2 3 Alex 50
Retain all values, all rows
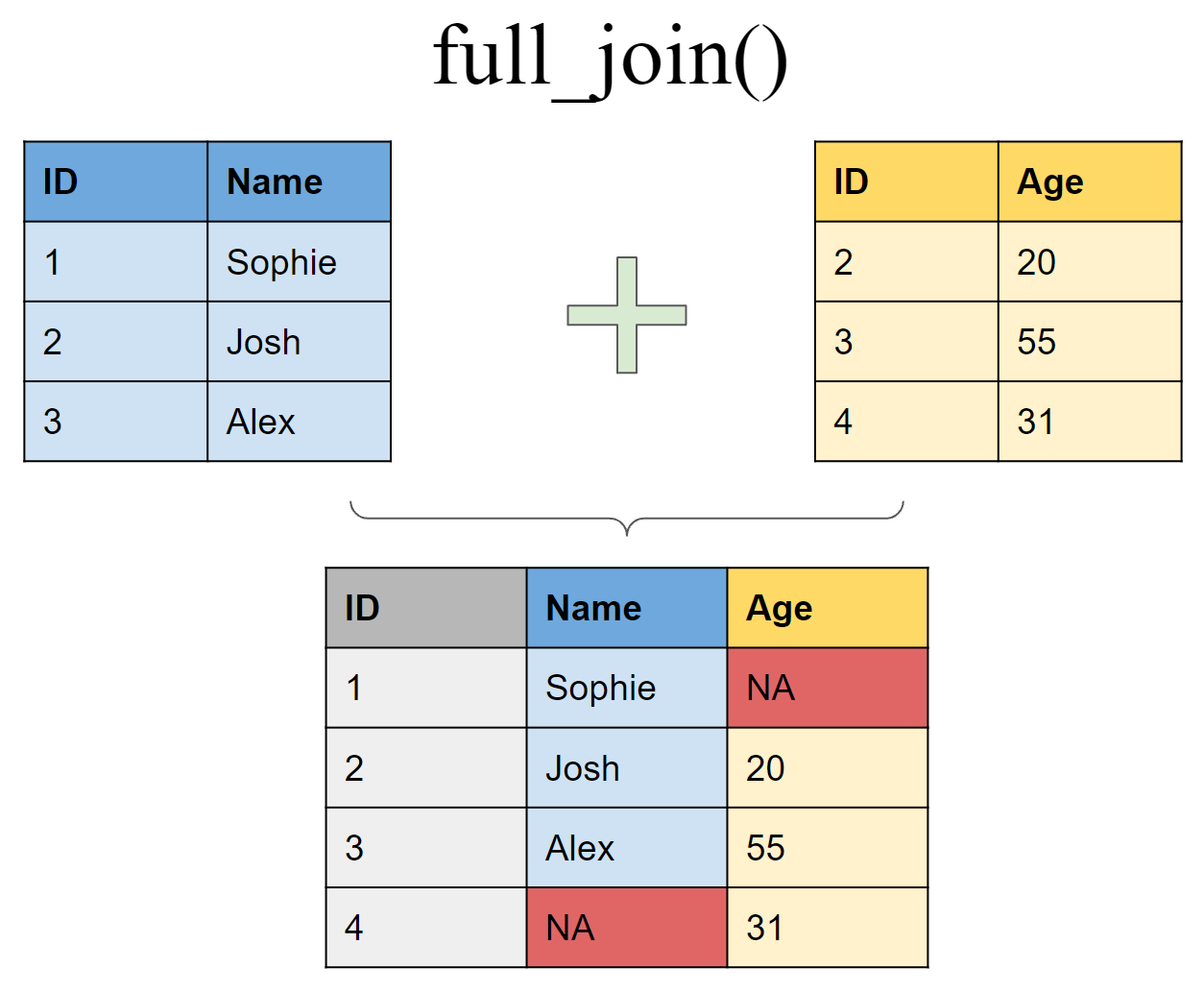
full_join(df1, df2, by = "ID")
## # A tibble: 4 x 3
## ID Name Age
## <int> <chr> <dbl>
## 1 1 Sophie NA
## 2 2 Josh 20
## 3 3 Alex 50
## 4 4 <NA> 31
Filtering joins
Retain all rows in df1 that have a match in df2
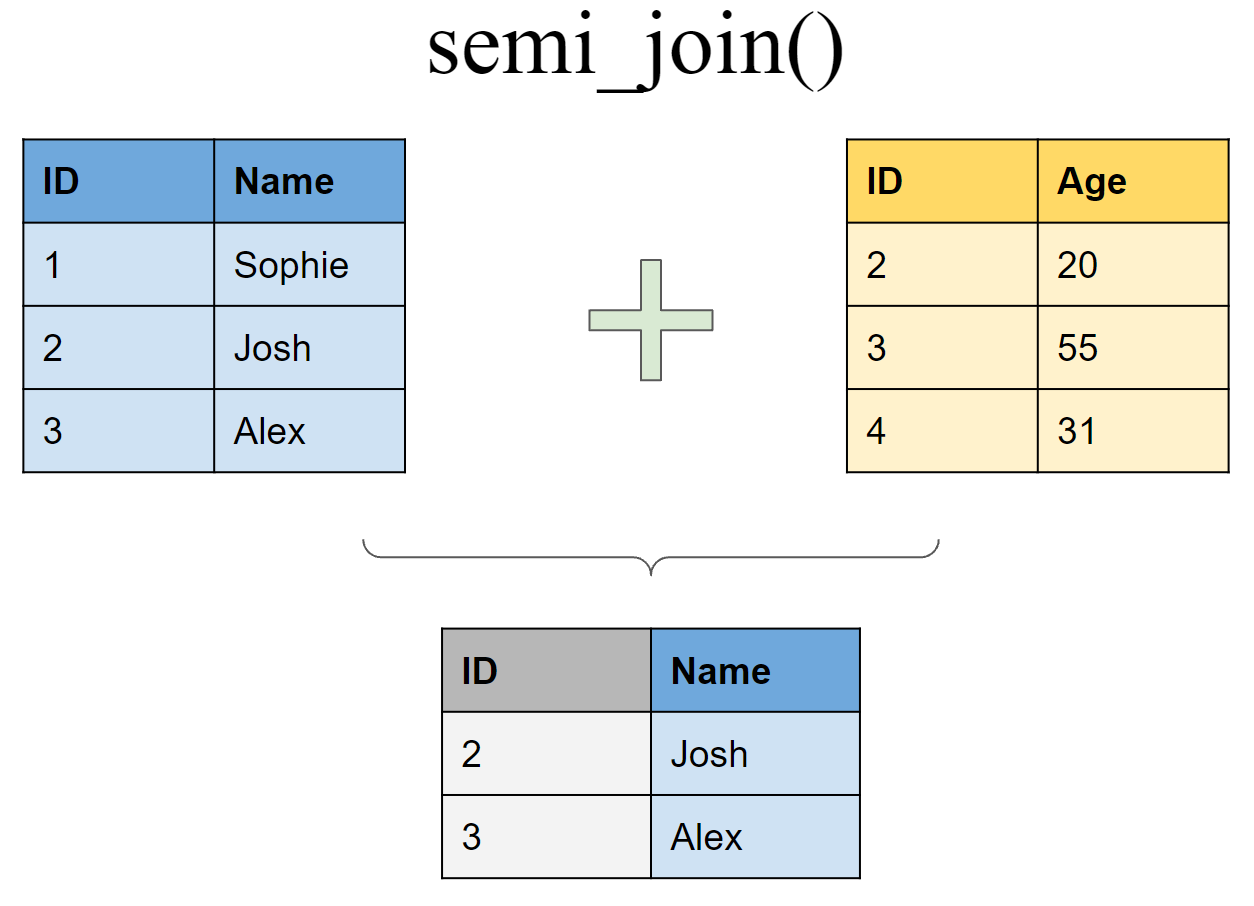
semi_join(df1, df2, by = "ID")
## # A tibble: 2 x 2
## ID Name
## <int> <chr>
## 1 2 Josh
## 2 3 Alex
Retain all rows in df1 that do not have a match in df2
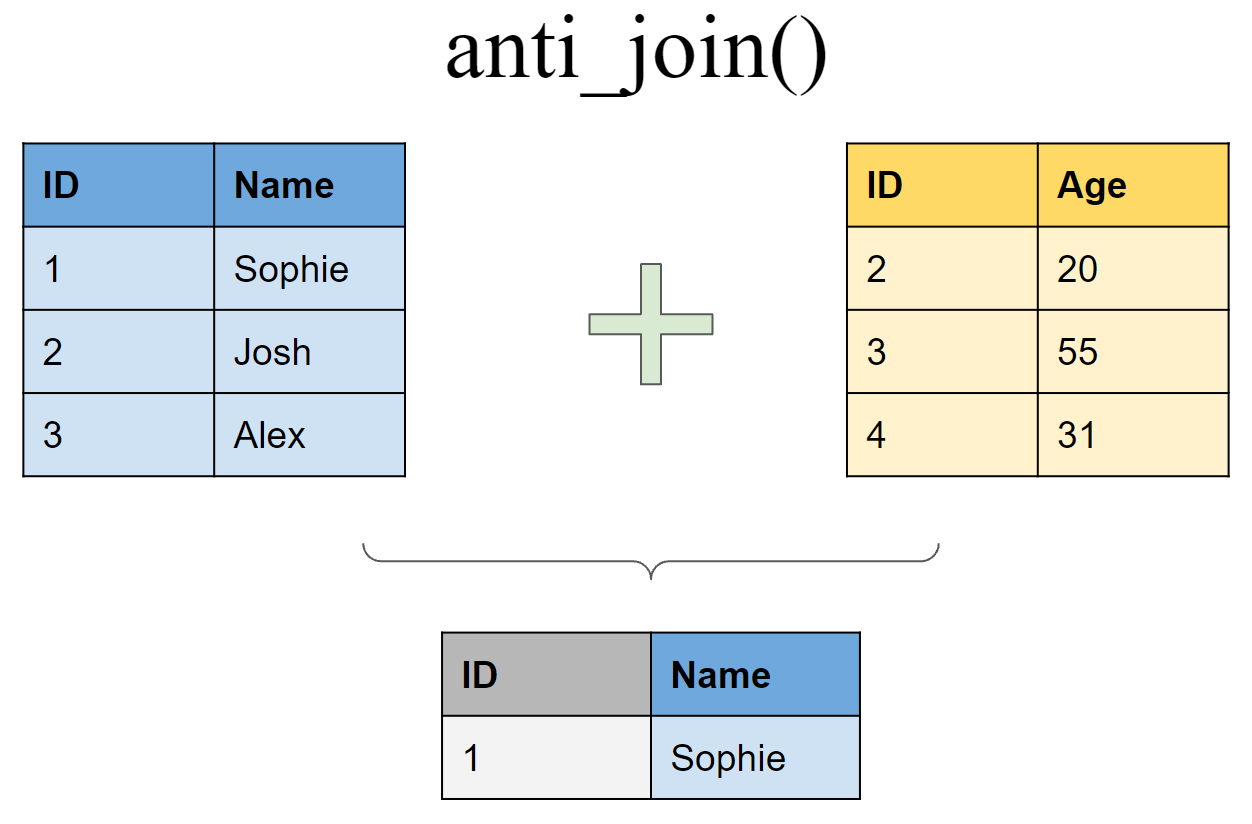
anti_join(df1, df2, by = "ID")
## # A tibble: 1 x 2
## ID Name
## <int> <chr>
## 1 1 Sophie
Binding
Append df2 to df1 as new rows
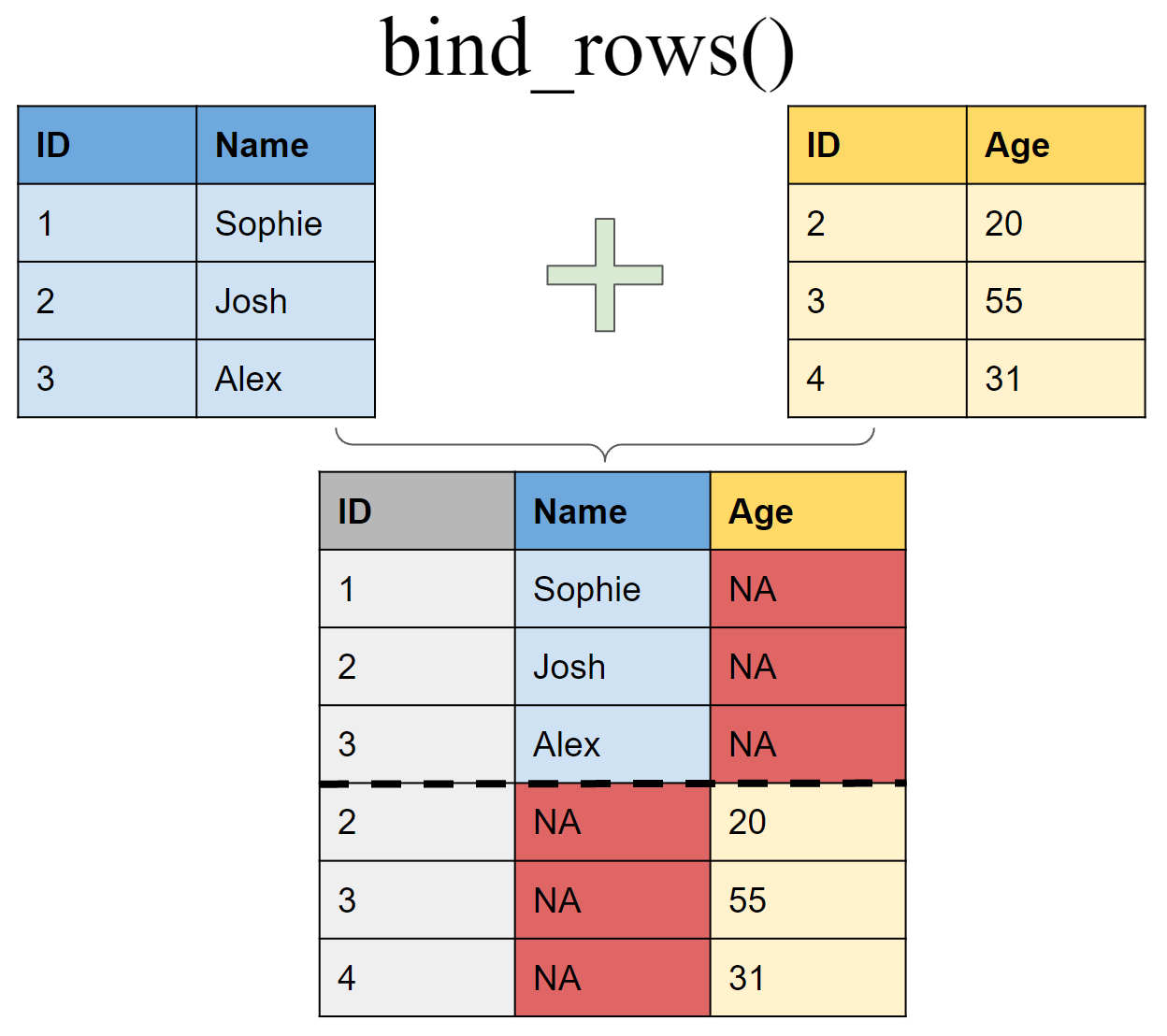
bind_rows(df1, df2)
## # A tibble: 6 x 3
## ID Name Age
## <int> <chr> <dbl>
## 1 1 Sophie NA
## 2 2 Josh NA
## 3 3 Alex NA
## 4 2 <NA> 20
## 5 3 <NA> 50
## 6 4 <NA> 31
Append df2 to df1 as new columns
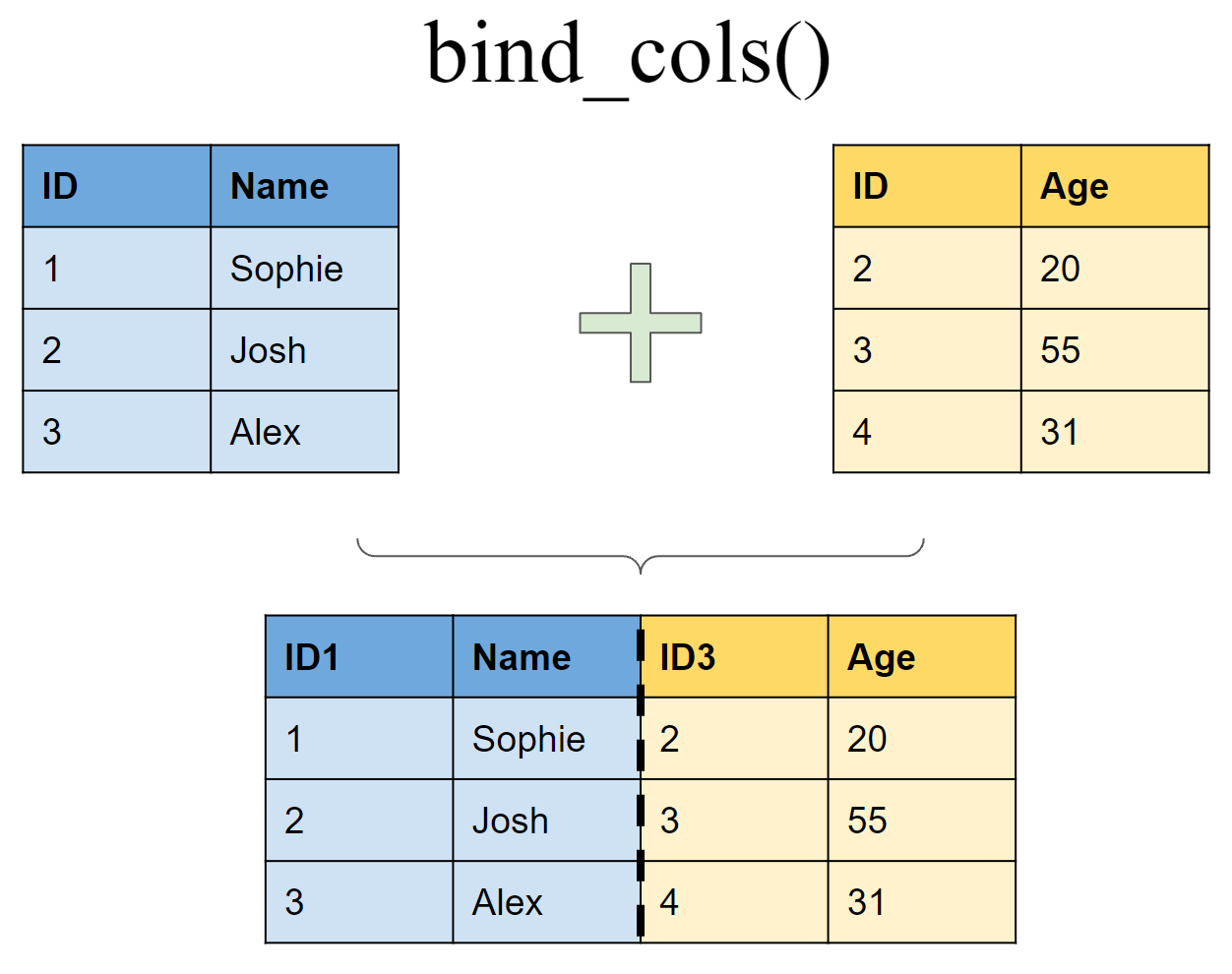
bind_cols(df1, df2)
## New names:
## * ID -> ID...1
## * ID -> ID...3
## # A tibble: 3 x 4
## ID...1 Name ID...3 Age
## <int> <chr> <int> <dbl>
## 1 1 Sophie 2 20
## 2 2 Josh 3 50
## 3 3 Alex 4 31
Joining multiple (>2) tibbles
Create a third tibble
df3 <- tibble(ID = 1:5,
Height = c(175,167,190,155,160))
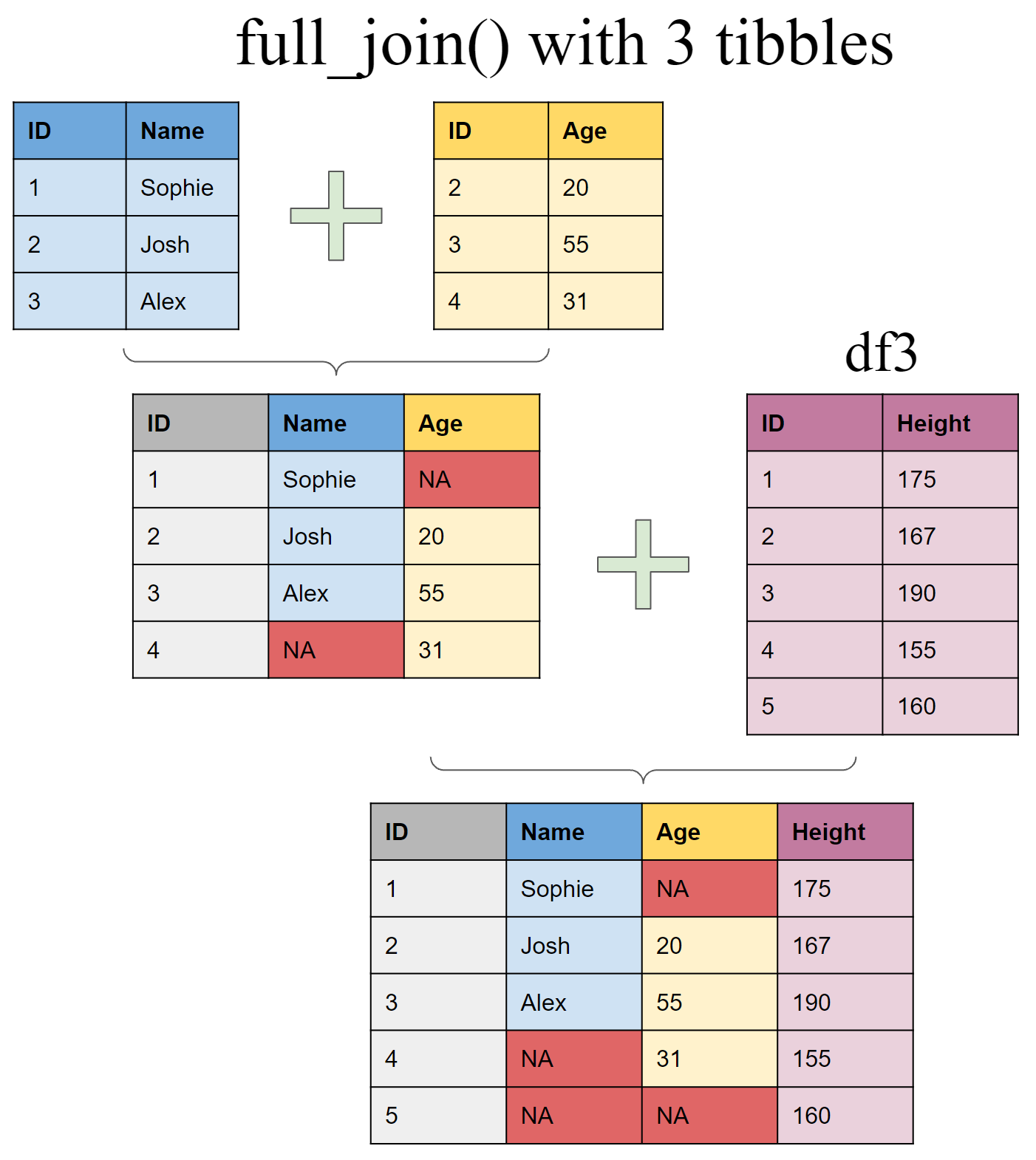
Use piping operator (%>%) to layer multiple join functions
full_join(df1, df2, by = "ID") %>%
full_join(df3, by = "ID")
## # A tibble: 5 x 4
## ID Name Age Height
## <int> <chr> <dbl> <dbl>
## 1 1 Sophie NA 175
## 2 2 Josh 20 167
## 3 3 Alex 50 190
## 4 4 <NA> 31 155
## 5 5 <NA> NA 160
Joining tibbles on multiple conditions
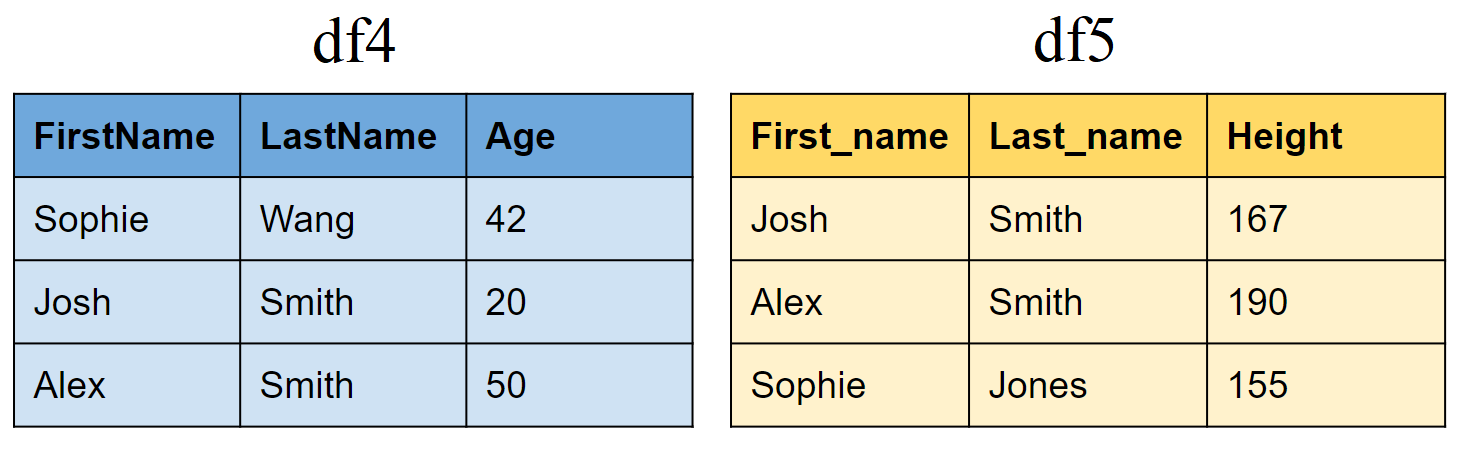
Create two new tibbles df4 and df5
df4 <- tibble(FirstName = c("Sophie", "Josh","Alex"),
LastName=c("Wang","Smith","Smith"),
Age = c(42,20,50))
df5 <- tibble(First_name = c("Josh","Alex","Sophie"),
Last_name=c("Smith","Smith","Jones"),
Height = c(167,190,155))
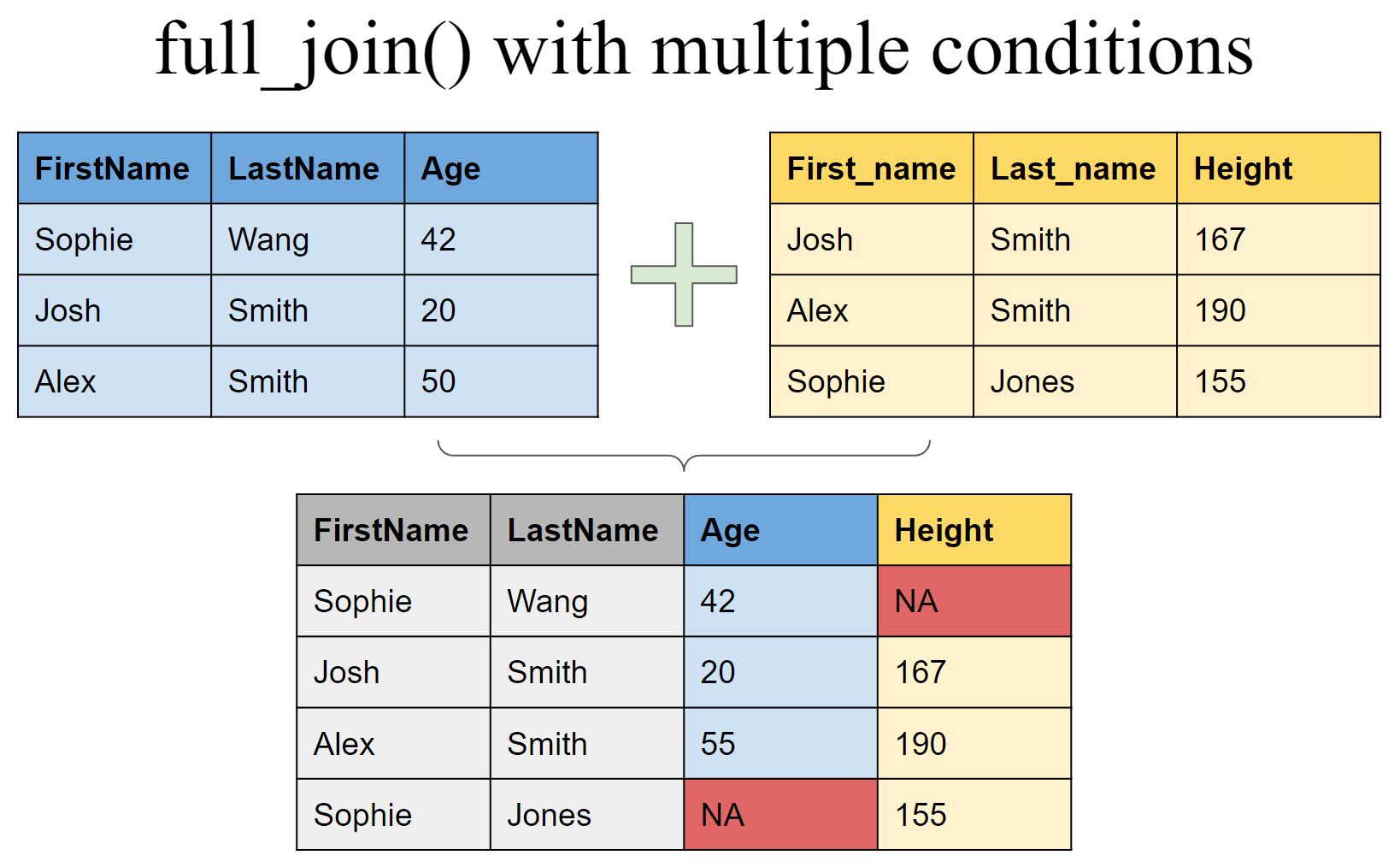
full_join(df4, df5, by = c("FirstName" = "First_name", "LastName" = "Last_name"))
## # A tibble: 4 x 4
## FirstName LastName Age Height
## <chr> <chr> <dbl> <dbl>
## 1 Sophie Wang 42 NA
## 2 Josh Smith 20 167
## 3 Alex Smith 50 190
## 4 Sophie Jones NA 155
What if you did not realize that multiple people shared the same Last Name?
full_join(df4, df5, by = c("LastName" = "Last_name"))
## # A tibble: 6 x 5
## FirstName LastName Age First_name Height
## <chr> <chr> <dbl> <chr> <dbl>
## 1 Sophie Wang 42 <NA> NA
## 2 Josh Smith 20 Josh 167
## 3 Josh Smith 20 Alex 190
## 4 Alex Smith 50 Josh 167
## 5 Alex Smith 50 Alex 190
## 6 <NA> Jones NA Sophie 155
What if you did not realize that multiple people shared the same First Name?
full_join(df4, df5, by = c("FirstName" = "First_name"))
## # A tibble: 3 x 5
## FirstName LastName Age Last_name Height
## <chr> <chr> <dbl> <chr> <dbl>
## 1 Sophie Wang 42 Jones 155
## 2 Josh Smith 20 Smith 167
## 3 Alex Smith 50 Smith 190
Set operations
Create sample tibbles
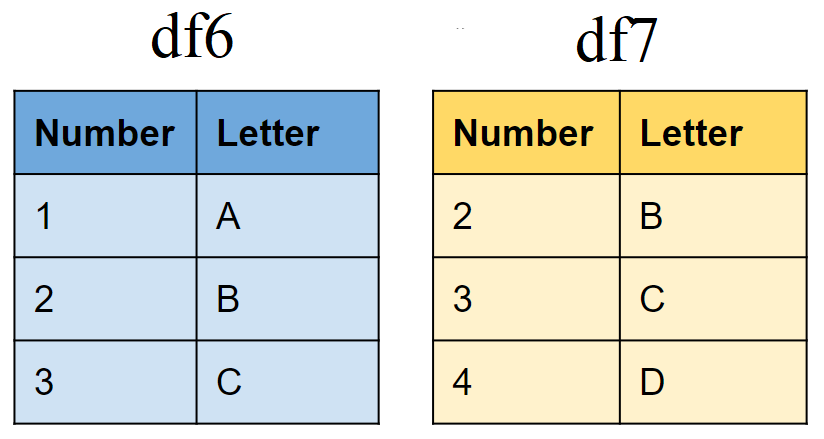
# First tibble
df6 <- tibble(Number = 1:3,
Letter = c("A", "B","C"))
# Second tibble
df7 <- tibble(Number = 2:4,
Letter = c("B","C","D"))
Include rows that appear in both tibbles
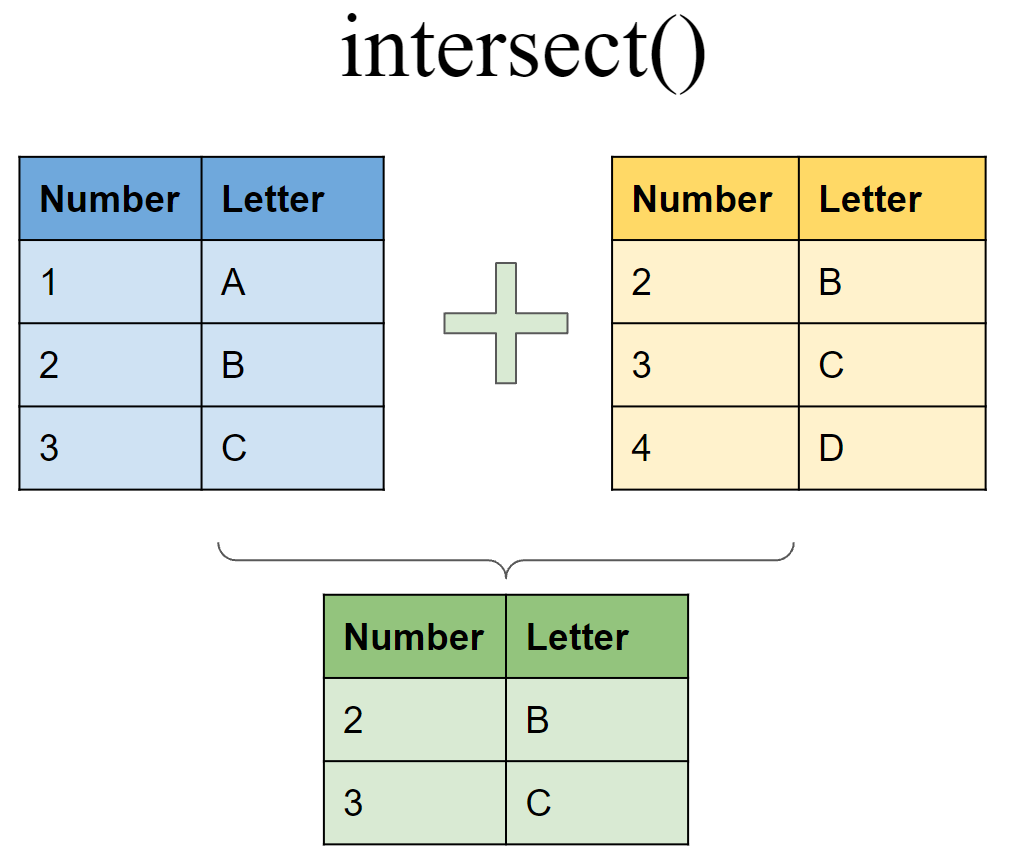
intersect(df6, df7)
## # A tibble: 2 x 2
## Number Letter
## <int> <chr>
## 1 2 B
## 2 3 C
Include rows that appear in either or both tibbles
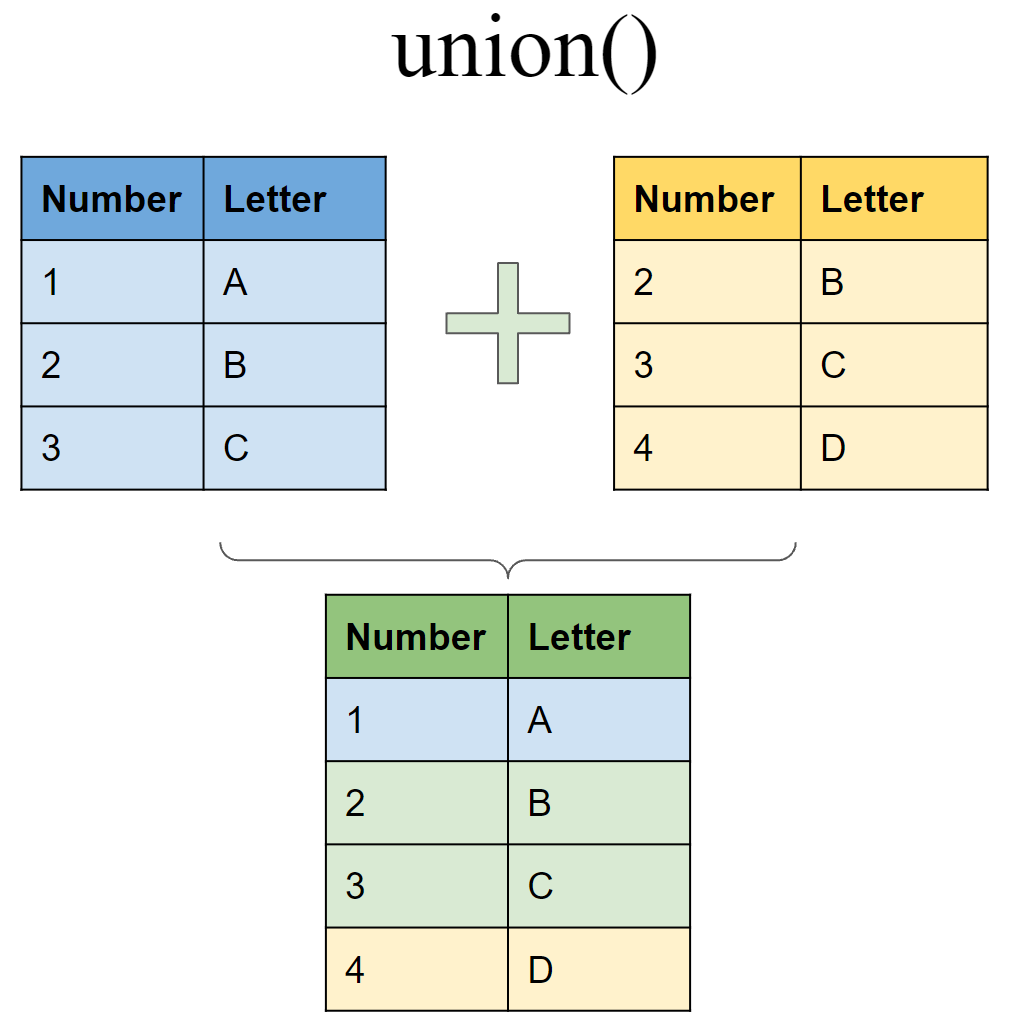
union(df6, df7)
## # A tibble: 4 x 2
## Number Letter
## <int> <chr>
## 1 1 A
## 2 2 B
## 3 3 C
## 4 4 D
Include rows that appear in one df but not another
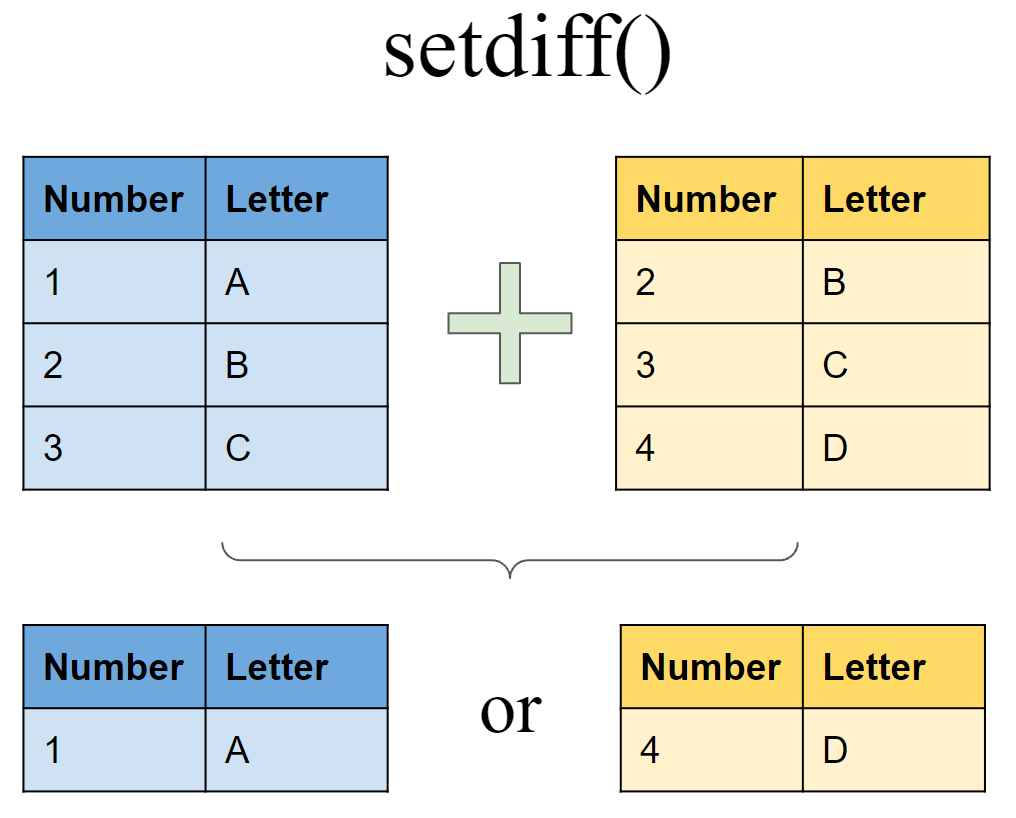
Include rows that appear in df6 but not in df7
setdiff(df6, df7)
## # A tibble: 1 x 2
## Number Letter
## <int> <chr>
## 1 1 A
Include rows that appear in df7 but not in df6
setdiff(df7, df6)
## # A tibble: 1 x 2
## Number Letter
## <int> <chr>
## 1 4 D
Joining tibbles with different types of variables
You can also join tibbles with sets of predictions:
set.seed(1)
x <- rnorm(5)
model1 <- tibble(x = x, yhat = 2.1 + 3.2 * x)
model2 <- tibble(x = x, yhat = 1.5 + 2.9 * x)
left_join(model1, model2, by = "x")
## # A tibble: 5 x 3
## x yhat.x yhat.y
## <dbl> <dbl> <dbl>
## 1 -0.626 0.0953 -0.317
## 2 0.184 2.69 2.03
## 3 -0.836 -0.574 -0.923
## 4 1.60 7.20 6.13
## 5 0.330 3.15 2.46
Demonstration with gapminder
Get an overview of gapminder data
glimpse(gapminder)
## Rows: 1,704
## Columns: 6
## $ country <fct> "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", ~
## $ continent <fct> Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, Asia, ~
## $ year <int> 1952, 1957, 1962, 1967, 1972, 1977, 1982, 1987, 1992, 1997, ~
## $ lifeExp <dbl> 28.801, 30.332, 31.997, 34.020, 36.088, 38.438, 39.854, 40.8~
## $ pop <int> 8425333, 9240934, 10267083, 11537966, 13079460, 14880372, 12~
## $ gdpPercap <dbl> 779.4453, 820.8530, 853.1007, 836.1971, 739.9811, 786.1134, ~
Part 1
Obtain additional information on countries from other open data sources
country_data <- read.csv(file = "https://raw.githubusercontent.com/open-numbers/ddf--gapminder--geo_entity_domain/master/ddf--entities--geo--country.csv")
glimpse(country_data)
## Rows: 273
## Columns: 33
## $ country <chr> "abkh", "afg", "akr_a_dhe", "ala", "alb", "dza"~
## $ gwid <chr> "i0", "i1", "i2", "i258", "i3", "i4", "i5", "i6~
## $ name <chr> "Abkhazia", "Afghanistan", "Akrotiri and Dhekel~
## $ world_6region <chr> "europe_central_asia", "south_asia", "europe_ce~
## $ income_groups <chr> "", "low_income", "", "", "upper_middle_income"~
## $ landlocked <chr> "", "landlocked", "coastline", "coastline", "co~
## $ g77_and_oecd_countries <chr> "others", "g77", "others", "others", "others", ~
## $ main_religion_2008 <chr> "", "muslim", "", "", "muslim", "muslim", "chri~
## $ gapminder_list <chr> "Abkhazia", "Afghanistan", "Akrotiri and Dhekel~
## $ alternative_1 <chr> "", "Islamic Republic of Afghanistan", "", "â\2~
## $ alternative_2 <chr> "", "", "", "", "", "", "", "", "", "", "", "",~
## $ alternative_3 <chr> "", "", "", "", "", "", "", "", "", "", "", "",~
## $ alternative_4_cdiac <chr> "", "Afghanistan", "", "", "Albania", "Algeria"~
## $ pandg <chr> "", "AFGHANISTAN", "", "", "ALBANIA", "ALGERIA"~
## $ god_id <chr> "GE-AB", "AF", "Akrotiri_Dhekelia", "AX", "AL",~
## $ alt_5 <chr> "", "", "", "", "", "", "", "", "", "", "", "",~
## $ upper_case_name <chr> "", "AFGHANISTAN", "", "AALAND ISLANDS", "ALBAN~
## $ iso3166_1_alpha2 <chr> "", "AF", "", "AX", "AL", "DZ", "AS", "AD", "AO~
## $ iso3166_1_alpha3 <chr> "", "AFG", "", "ALA", "ALB", "DZA", "ASM", "AND~
## $ iso3166_1_numeric <int> NA, 4, NA, 248, 8, 12, 16, 20, 24, 660, 10, 28,~
## $ iso3166_2 <chr> "", "", "", "", "", "", "", "", "", "", "", "",~
## $ unicode_region_subtag <chr> "", "AF", "", "AX", "AL", "DZ", "AS", "AD", "AO~
## $ arb1 <chr> "", "", "", "", "", "", "", "", "", "", "", "",~
## $ arb2 <chr> "", "", "", "", "", "", "", "", "", "", "", "",~
## $ arb3 <chr> "", "", "", "", "", "", "", "", "", "", "", "",~
## $ arb4 <chr> "", "", "", "", "", "", "", "", "", "", "", "",~
## $ arb5 <chr> "", "", "", "", "", "", "", "", "", "", "", "",~
## $ arb6 <chr> "", "", "", "", "", "", "", "", "", "", "", "",~
## $ is..country <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,~
## $ un_state <lgl> FALSE, TRUE, FALSE, FALSE, TRUE, TRUE, FALSE, T~
## $ world_4region <chr> "europe", "asia", "europe", "europe", "europe",~
## $ latitude <dbl> NA, 33.00000, NA, 60.25000, 41.00000, 28.00000,~
## $ longitude <dbl> NA, 66.00000, NA, 20.00000, 20.00000, 3.00000, ~
Narrow down information to income groups, OECD status, and religion
country_data <- country_data %>%
select(name, income_groups, g77_and_oecd_countries, main_religion_2008)
# Check data structure
glimpse(country_data)
## Rows: 273
## Columns: 4
## $ name <chr> "Abkhazia", "Afghanistan", "Akrotiri and Dhekel~
## $ income_groups <chr> "", "low_income", "", "", "upper_middle_income"~
## $ g77_and_oecd_countries <chr> "others", "g77", "others", "others", "others", ~
## $ main_religion_2008 <chr> "", "muslim", "", "", "muslim", "muslim", "chri~
Count how many unique country names are in gapminder and country_data
nlevels(gapminder$country)
## [1] 142
nlevels(as.factor(country_data$name))
## [1] 273
Merge gapminder and country_data using left_join()
gapminder_extended <- left_join(gapminder, country_data, by=c("country"="name"))
head(gapminder_extended)
## # A tibble: 6 x 9
## country continent year lifeExp pop gdpPercap income_groups
## <chr> <fct> <int> <dbl> <int> <dbl> <chr>
## 1 Afghanistan Asia 1952 28.8 8425333 779. low_income
## 2 Afghanistan Asia 1957 30.3 9240934 821. low_income
## 3 Afghanistan Asia 1962 32.0 10267083 853. low_income
## 4 Afghanistan Asia 1967 34.0 11537966 836. low_income
## 5 Afghanistan Asia 1972 36.1 13079460 740. low_income
## 6 Afghanistan Asia 1977 38.4 14880372 786. low_income
## # ... with 2 more variables: g77_and_oecd_countries <chr>,
## # main_religion_2008 <chr>
Note:: left_join() is probably the most useful and the most used join. It is often used when you want to expand your existing dataset with new variables from other sources.
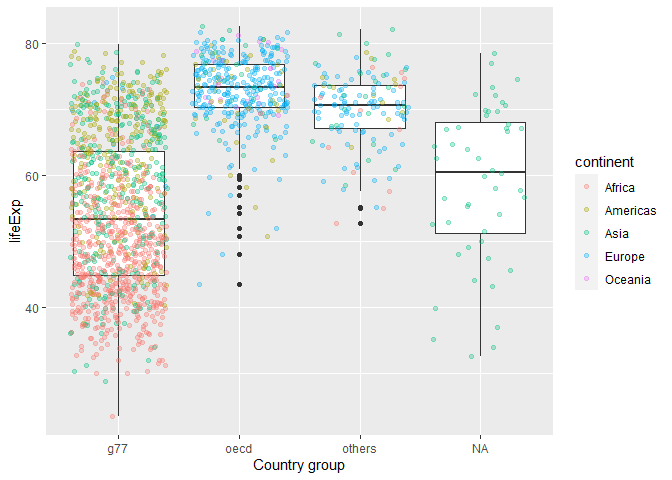
Compare lifeExp for OECD, G77, and other countries
gapminder_extended %>%
ggplot(aes(x=g77_and_oecd_countries,y=lifeExp))+
geom_boxplot()+
geom_jitter(aes(color=continent), alpha=0.3)+
labs(x="Country group")
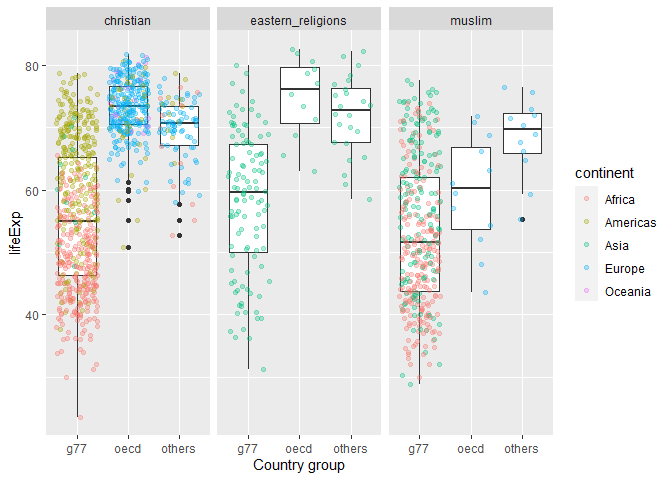
Compare lifeExp for OECD, G77, and other countries by most common religion
gapminder_extended %>%
filter(main_religion_2008 %in% c("christian","eastern_religions","muslim")) %>%
ggplot(aes(x=g77_and_oecd_countries,y=lifeExp))+
geom_boxplot()+
geom_jitter(aes(color=continent), alpha=0.3)+
labs(x="Country group")+
facet_wrap(~main_religion_2008)
Part 2
Gapminder data is only available from 1952 to 2007. What if we wanted to examine data after 2007 as well as population projections?
Download population size estimates by country from 1800 to 2100
population <- gsheet2tbl("https://docs.google.com/spreadsheets/d/14_suWY8fCPEXV0MH7ZQMZ-KndzMVsSsA5HdR-7WqAC0/edit#gid=176703676")
See what population data looks like
glimpse(population)
## Rows: 59,297
## Columns: 4
## $ geo <chr> "afg", "afg", "afg", "afg", "afg", "afg", "afg", "afg", "af~
## $ name <chr> "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan",~
## $ time <dbl> 1800, 1801, 1802, 1803, 1804, 1805, 1806, 1807, 1808, 1809,~
## $ Population <dbl> 3280000, 3280000, 3280000, 3280000, 3280000, 3280000, 32800~
Only retain population estimates after 2007, rename variables to match gapminder variable names
population <- population %>%
filter(time>2007) %>%
rename(year=time, country=name, pop=Population) %>%
select(-geo)
Add continent data to population from gapminder
# create a data frame listing continent for every country
continent <- gapminder %>%
select(country, continent) %>%
distinct()
# add continent data to population data frame
population <- left_join(population, continent, by = "country")
# see how many countries are missing continent data by continent
population %>%
group_by(year) %>%
summarise(missing_continent = sum(is.na(continent)))
## # A tibble: 93 x 2
## year missing_continent
## <dbl> <int>
## 1 2008 61
## 2 2009 61
## 3 2010 61
## 4 2011 61
## 5 2012 61
## 6 2013 61
## 7 2014 61
## 8 2015 61
## 9 2016 61
## 10 2017 61
## # ... with 83 more rows
Use bind_rows() to stack population below gapminder
gapminder_pop <- bind_rows(gapminder, population) %>%
arrange(country,year)
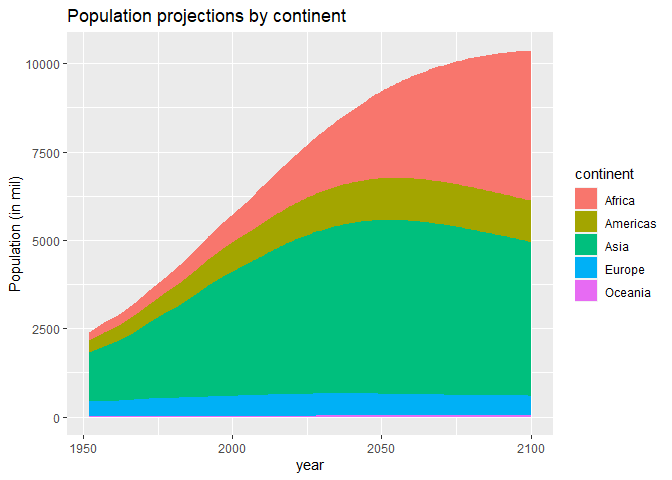
Visualize trends in population growth by continent
gapminder_pop %>%
filter(!is.na(continent)) %>%
group_by(continent, year) %>%
summarise(pop=sum(pop)/1000000) %>%
ggplot(aes(x=year, y=pop, fill=continent))+
geom_area()+
labs(title="Population projections by continent",
y="Population (in mil)")
## `summarise()` has grouped output by 'continent'. You can override using the `.groups` argument.
Attributions
Written by Albina Gibadullina with input from Vincenzo Coia.